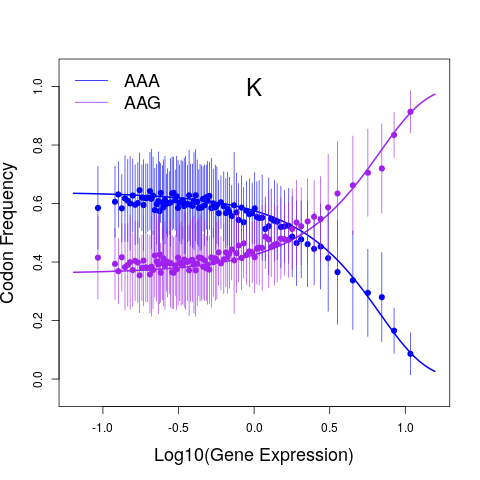
Synonymous codons are not used used at equal frequencies, a phenomenon known as codon usage bias (CUB) that is observed across the major domains of life. CUB arises due to a combination of adaptive (e.g., natural selection for fast translation) and non-adaptive evolutionary processes (e.g., biases in DNA mutations), but the relative importance of these different processes remains controversial. Furthermore, CUB often varies between even closely-related species, but the evolutionary causes for such sudden shifts in CUB remains unknown. Analyses of CUB often rely on heuristic measures of CUB that are difficult to interpret in explicitly evolutionary terms. By viewing CUB through an explicitly evolutionary lens using population genetics models of coding sequence evolution, I derived new insights into the roles of adaptive and non-adpative evolutionary processes shaping codon usage. Going forward, I will develop new models of CUB evolution that explicitly link biophysical models of molecular processes to models of evolutionary fitness. Using these biophysical-evolutionary models, I will derive new insights into the role of different evolutionary processes in shaping CUB and how these different processes interact and interply to shape proteome-wide CUB.
Shifts in gene expression are a major contributor to phenotypic variation within and across species, Gene expression is the result of multiple regulatory elements (REs) that span transcription to post-translation, leading to a complex interplay in the evolution of different REs. Phylogenetic studies of gene expression have emerged as a powerful approach to test if variation in gene expression across species is consistent with non-adaptive or adaptive evolution. Although useful, phylogenetic studies of are gene expression are often detached from the evolution of the underlying gene regulation. Building off my expertise in evolutionary genetics, transcriptomics, and proteomics, I study gene expression evolution at multiple regulatory levels using a combination of bioinformatics and phylogenetic approaches.