Synonymous codons are not used used at equal frequencies, a phenomenon known as codon usage bias (CUB) that is observed across the major domains of life. CUB arises due to a combination of adaptive (e.g., natural selection for fast translation) and non-adaptive evolutionary processes (e.g., biases in DNA mutations), but the relative importance of these different processes remains controversial. Furthermore, CUB often varies between even closely-related species, but the evolutionary causes for such sudden shifts in CUB remains unknown. Analyses of CUB often rely on heuristic measures of CUB that are difficult to interpret in explicitly evolutionary terms. By viewing CUB through an explicitly evolutionary lens using population genetics models of coding sequence evolution, I derived new insights into the roles of adaptive and non-adpative evolutionary processes shaping codon usage. Going forward, I will develop new models of CUB evolution that explicitly link biophysical models of molecular processes to models of evolutionary fitness. Using these biophysical-evolutionary models, I will derive new insights into the role of different evolutionary processes in shaping CUB and how these different processes interact and interply to shape proteome-wide CUB.
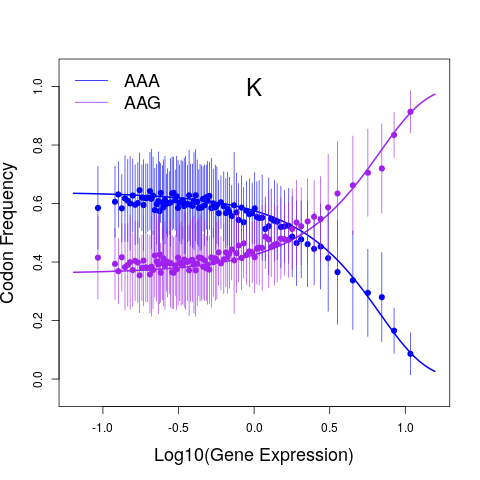
Macroevolutionary changes in natural selection on codon usage reflect evolution of the tRNA pool across a budding yeast subphylum Cope and Shah 2025. PNAS
The Importance of Nonsense Errors: Estimating the Rate and Implications of Drop-Off Errors during Protein Synthesis Cope et al. 2024. bioRxiv
Evolutionary principles underpinning codon usage bias: patterns, functions, and mechanisms Cope et al. 2024. EcoEvoRxiv
Re-examining Correlations Between Synonymous Codon Usage and Protein Bond Angles in Escherichia coli Akeju and Cope, Gen. Biol. and Evol.
Intragenomic variation in non-adaptive nucleotide biases causes underestimation of selection on synonymous codon usage Cope and Shah 2022. Plos Genetics.
Quantifying shifts in natural selection on codon usage between protein regions: a population genetics approach Cope and Gilchrist 2022. BMC Genomics.
Quantifying codon usage in signal peptides: Gene expression and amino acid usage explain apparent selection for inefficient codons Cope et al. 2018. Biochimica et Biophysica Acta (BBA) - Biomembranes.
AnaCoDa: analyzing codon data with Bayesian mixture models Landerer et al. 2018. Bioinformatics.